The Image Processing Environment
The component provides the interactive display and editing capabilities of the package. Datasets in our problem domain fall into three categories:
- Volumes: 3D datasets from MRI examinations to be displayed as images (slices or 3D projections),
- Signals: 2D (channel vs. time) datasets from EEG and MEG measurements usually displayed as curves, and
- Geometrical Objects: Surfaces and volumetric meshes computed from 3D datasets.
Viewers (for data display) and editors (for data modification) are provided for all three categories. Cross-category data can be displayed together: a geometrical object (i.e., the surface of a tumor) is rendered inside a volumetric dataset of the brain; the electric potential recorded on the scalp is mapped as a color-coded texture onto the geometrically defined surface of the head. Viewers can exchange information: cursor movements or contrast adjustments in an orthogonal viewer are propagated to the other projections; selection of a time point in a signal viewer maps the activity onto the head\'s surface display; selecting a position on this surface highlights the nearest electrode trace in the signal viewer. Editors provide functions for modifying or annotating data, e.g., expert segmentation of brain structures. The state of the kernel including all viewers may be saved into a file as a "workspace".
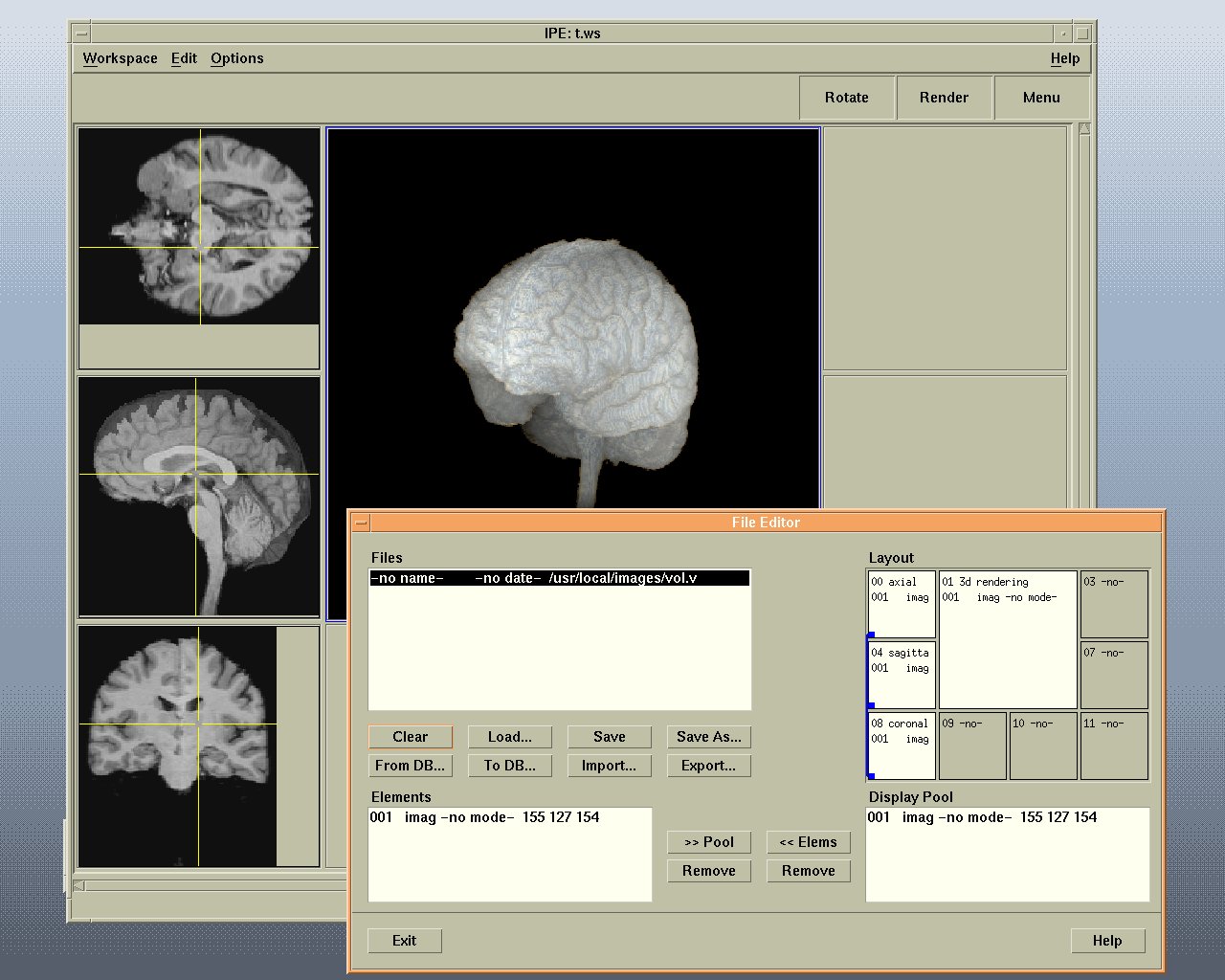 |
Example BRIAN workspace showing two geometrical viewer windows (bottom row), a volume renderer (top right) and three orthogonal viewers (top left). All viewers are connected via links, so that cursor movements are coordinated.
